
During the unfolding event (left to right), the solvent becomes more ordered, which is unfavorable. Q: I have generated an ESPript figure.\( \newcommand\)=1. This latter must correspond to the last sequence of your alignment. In this goal, switch to ADV mode.Ī BOTTOM secondary structures section is now displayed on the form, where you can put a structure file as input. Notice that you can insert secondary structure elements at the bottom of an alignment. Example: 10 all (means that the sequence order of your alignment is changed in order to represent sequence number 10 first, then all the others). In the Defining groups section of the ESPript form, you can change the order of the sequences.Manually change the order of the sequence in your alignment file: put the sequence line corresponding to the secondary structure file first.The Secondary structure depiction section refers to the secondary structure elements to be displayed at the top of the sequence block.īy default, ESPript supposes that the secondary structure file corresponds to the first sequence in the alignment. A click on the RESULT button achieves nothing. Q: When I submit a job, I cannot find the results anywhere. You can follow the ESPript tutorials to get familiar with such process.Īlternately, you can have a look at ENDscript ( ) which can help you, in certain cases, to produce complex ESPript figures. Finally, as there is no possibility to have PDB or DSSP files containing multiple structures, you need to add one structure file per sequence. Please take due note that ESPript is not able to predict protein secondary structures : it only uses experimentally determined structures.

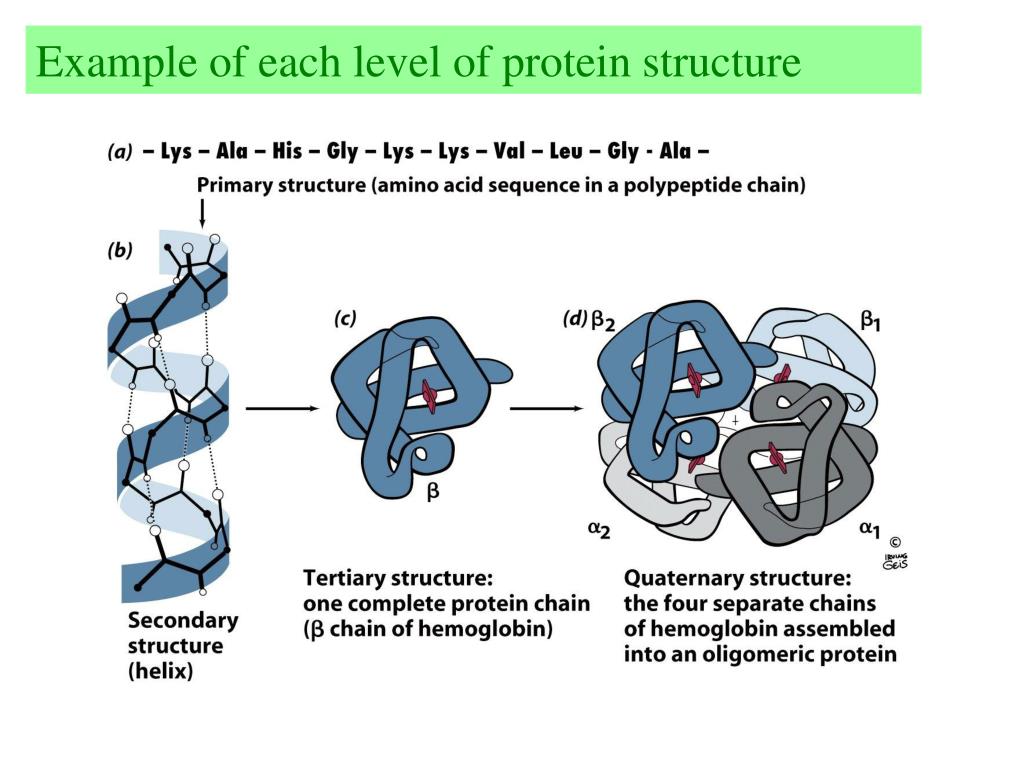
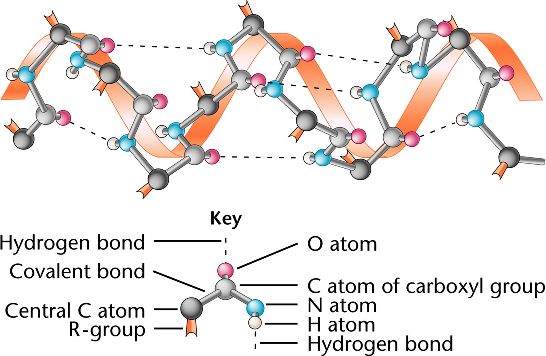
If a 3D structure corresponding to a sequence is known (in PDB or DSSP format only), you can add it to the figure.
Then, you need to upload this file in ESPript to produce a basic multiple alignment figure. As an example, you can perform such alignment with online servers like EMBL-EBI Clustal Omega or ClustalW2. The starting point of an ESPript figure is a protein multiple sequence alignment file in Clustal, FASTA, MultAlin, or ProDom format.


 0 kommentar(er)
0 kommentar(er)
